The modular framework in TinkerCell is designed to support practically any format that can be loaded into TinkerCell. As a consequence, Antimony and SBML files can be used as modules. In the schreenshot below, a TinkerCell oscillator module is modeled using an Antimony script. The modular framework maps variables in the Antimony script to variables in the conceptual model.
TinkerCell is a Computer-Aided Design software tool for Synthetic Biology that promotes collaboration through its plugin interface. This blog is used to keep notes on updates to the project.
Tuesday, September 13, 2011
Friday, September 2, 2011
Same network, different behaviors
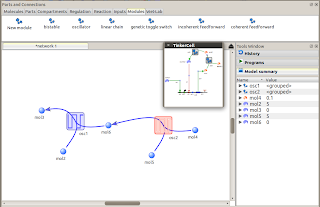
Coupled oscillator modules
TinkerCell's modular framework allows users to perform experiments such as the one shown below. Here, two oscillators are connected to each other. Each oscillator *can* take multiple models. This is the benefit of using modular design -- the internal details can be swapped without affecting the conceptual overall design, i.e. two coupled oscillators. One of the TinkerCell Python plugins allows users to test all possible oscillator combinations. Currently, only two oscillator designs are available (you can add more and submit to the repository if you want!). So, the total number of combinations are 4, which are the 4 graphs shown below. Notice that in one of the designs (top left), one oscillator somewhat controls the frequency of the other. By tuning parameters further, this design can actually be used to create an oscillator where the frequency is also oscillating.
Subscribe to:
Comments (Atom)


