
TinkerCell is a Computer-Aided Design software tool for Synthetic Biology that promotes collaboration through its plugin interface. This blog is used to keep notes on updates to the project.
Sunday, January 30, 2011
Sticky ends
See snapshots of the new "Sticky ends" part. It is not very useful for the purpose of modeling. But if TinkerCell should include plugins that do some assembly-related operations, perhaps this can be useful...maybe


Saturday, January 29, 2011
Sharing models and code made easy
New Python and Octave code can be submitted to the repository directly from TinkerCell. The "Save" button in TinkerCell's coding window will do the work for the user. Basically, it will generate the SVN command for uploading to the repository (the user must have subversion installed). The "tinkercelluser" sourceforge account is used for the upload process.

It is also simple for a user to submit models to the repository:
File -> Export -> Share model

The above dialog appears, asking the user for the name and family of the model. The interface at present does not allow the user to create new families of models; that will require modifying the modules.xml file and populating (correctly) the participant types and roles, which might be a bit too much for most users. So adding new families is kept as something that only more advanced users would do (and they would directly edit the xml file)

It is also simple for a user to submit models to the repository:
File -> Export -> Share model

The above dialog appears, asking the user for the name and family of the model. The interface at present does not allow the user to create new families of models; that will require modifying the modules.xml file and populating (correctly) the participant types and roles, which might be a bit too much for most users. So adding new families is kept as something that only more advanced users would do (and they would directly edit the xml file)
Graphical tooltips for sub-models and functions
The decorator icon is also modified so that it is consistently the same icon for all connections that have a hidden model inside. This is due to confusions caused by the inconsistent meanings of the other decorators.

In the next screenshot, the tool-tip window displays the function that is used to assign the value to the repressor binding site. This is important because TinkerCell allows different modeling strategies, so the user (quickly) should know when an assignment rule is used.

PDF export finally works
The exported PDF (from "screenshot" button) is clearly using vector graphics and not pixels.

Friday, January 28, 2011
Small change to assignment rules
And parts on plasmid look better now (side bars become invisible)

Tuesday, January 18, 2011
Automatically create and upload installer
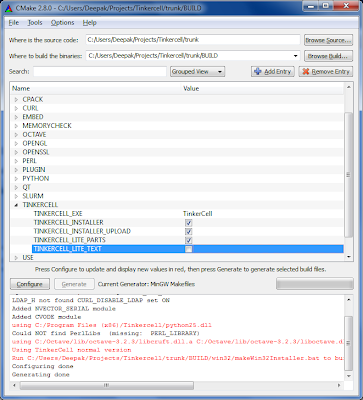
In Windows and Mac, it is a bit time consuming to create the TinkerCell installer and upload it to Sourceforge.
As a solution, I created a simple CMake-generated script that does the work. Different scripts are generated based on the operating system. The Windows and Linux scripts build the program and upload it to sourceforge. Linux script can also upload the source code. The Mac script does not upload yet, but it does generate the image file (which is most of the work). For developers who want to build TinkerCell on their own machines, new instructions are available in the README.txt file included with the code. Here is a summary:
1. (windows) Install Inno setup, ISTools, and WinSCP.. and CMake of course
2. Download TinkerCell source code
3. Run CMake GUI (use the "grouped" view for convenience)
4. Set the TinkerCell source folder (/trunk) and the build folder (/trunk/BUILD)
5. Select TinkerCell lite versions if desired (under the TinkerCell group)
6. Run "Configure" and "Generate". Look for the message in red, which will tell you where the installation script is created
7. Run the installation script. It will create the program, the installer, and upload it!
Steps 1-6 only need to be run the first time when setting up CMake
For Mac...all the steps are the same, except for #7
7. Run the create_bundled_app.sh script in the BUILD/bin folder
For Linux, there is the additional option of selecting 64-bit or 32-bit OS. The is also the option of uploading the source along with the binaries...
7. Run upload.sh
Monday, January 17, 2011
TinkerCell "lite" revisited
Sunday, January 16, 2011
Integrated COPASI and sliders

The COPASI C++ API has been integrated into TinkerCell. Simulation functions, MCA functions, and steady state analysis function have been added. The image above shows a plot of the Eigenvalues with sliders to visualize how the Eigenvalues change due to change in parameters.
A COPASI C API has been created. The API uses Hash tables for quick look-up, thus adding the convenience of name-based look-up. See copasi_api.h for the functions.
Subscribe to:
Posts (Atom)

